Register
To register to the event, please click on the button below to fill the form. The deadline for registration is Friday 27 November.
Registration is now closed.
Program
Please find bellow the program of the Brainhack event (timezone Paris UTC+1). If you want to add the following schedule to your own personal Google agenda, click on the "Save the dates" button and then click on the "+ Google agenda" located at the bottom right of your screen.
2nd December
| 09h00-10h15 | Introduction to the BrainHack Marseille
|
|---|---|
| 10h15-12h30 | Hacking |
| 12h30-14h00 | Break |
| 14h00-15h00 |
Software demonstration
|
| 15h00-17h00 | Hacking |
3rd December
| 09h00-11h30 | Hacking |
|---|---|
| 11h30-12h30 | Unconference - Cyril Pernet
Recommendations for reproducible EEG and MEG research (from the OHBM Cobidas committee), (Pernet et al. Nature Neuro 2020) |
| 12h30-14h00 | Break |
| 14h00-16h00 | Hacking |
| 16h00-17h00 | Unconference - Thomas Brochier & Julia Sprenger
An open electrophysiological dataset recorded in non-human primates: acquisition, curation and exploitation using open source tools (odML, neo, Elephant), (Brochier et al. Sci Data 2018) |
4th December
| 09h00-12h30 | Hacking |
|---|---|
| 12h30-14h00 | Break |
| 14h00-15h00 | Unconference - Guillaume Dumas
Hands on experiments about reproducible analyses, (Kim et al. GigaScience 2018) |
| 15h00-16h00 | Hacking |
| 16h00-17h00 | Wrapping up about the projects with everybody, Ending and virtual beer ! |
Projects
Here you can find all the informations about the event projects.
If you want to submit a project you should follow the link, fill the form, and open a github issue. Projects can be anything you'd like to work on during the event with other people (coding, discussing a procedure with coworkers, brainstorming about a new idea), as long as you're ready to minimally organize this!
Optimization and GPU porting of information flow implementation
by Etienne Combrisson & Ruggero Basanisi
The following points are going to be addressed in this project:
- Code a GPU implementation of the conditional mutual-information
- Include a switch for the user to specify if a CPU or a GPU implementations are going to be used when computing the univariate information flow
- GPU porting of the multivariate information flow measure (which is also going to use the GPU version of scikit-learn)
Required skills
This is a python based project, a github link will be provided to collaborate.
eCOBIDAS: a webApp to that writes your methods section for you
by Rémi Gau
You can find several "good first issues" here but during this event I would like to focus on:
- Making sure the documenation is understandable and constitute good on-board and How-To materieal
- Create some prototype script / toy example that can take the output of the app and generate a method section
Required skills
The project has many moving parts so there is a certain level of flexibility in terms of skills needed.
DIGLaB : DIGital Lab Book
by Fred Barthelemy, Sylvain Takerkart & Julia Sprenger
- Can be read by automatic process for storage of comments or metadata
- Can help to automatize the writing of information in a click
- Can help to adopt future INT (or other) standards easily (e.g. filenames)
- Can be printed in case of need
- Avoid fastidious decoding of someone else's handwriting
- Present the current version
- Get feedback from future users
- Provide personalized versions to the users that want to use it
- Develop a tool for automatice metadata collection from saved DIGLaBs
Required skills
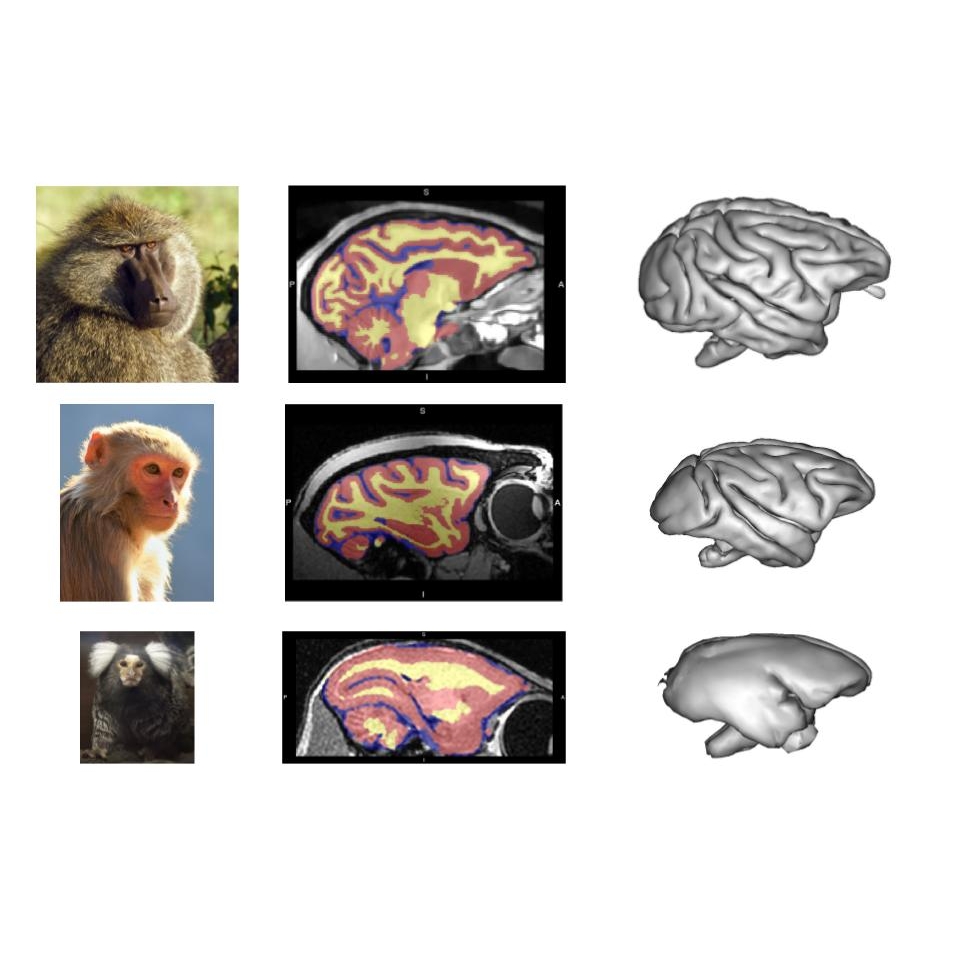
Macapype: segmentation to mesh pipeline
by David Meunier, Bastien Cagna & Kep Kee Loh
Link to the mattermost users forum (requires a framateam account).
In this brainhack, we would like to expand the existing pipelines of Macapype to allow the generation of surface meshes following the segmentation of the MR images. So far, at the INT, macapype users have been performing the above process (i.e. surface mesh generation) by first, using a set of customised scripts to import Macapype-generated segmentations into Brainvisa, and second, to manually (point-and-click) generate surface meshes via the Morphologist Toolbox in Brainvisa. We aim to incorporate these two steps as an additional module in Macapype, which will enable the generation of surface meshes directly from the segmentations produced by Macapype. The final result would be a powerful pipeline that takes raw PNH structural MR scans as input, and generating tissue segmentation masks and surface meshes as outputs.
Required skills
Other related softwares can be used for this project, such as BrainVISA, FreeSurfer and Nipype. Anyone that has PNH data to process is welcome.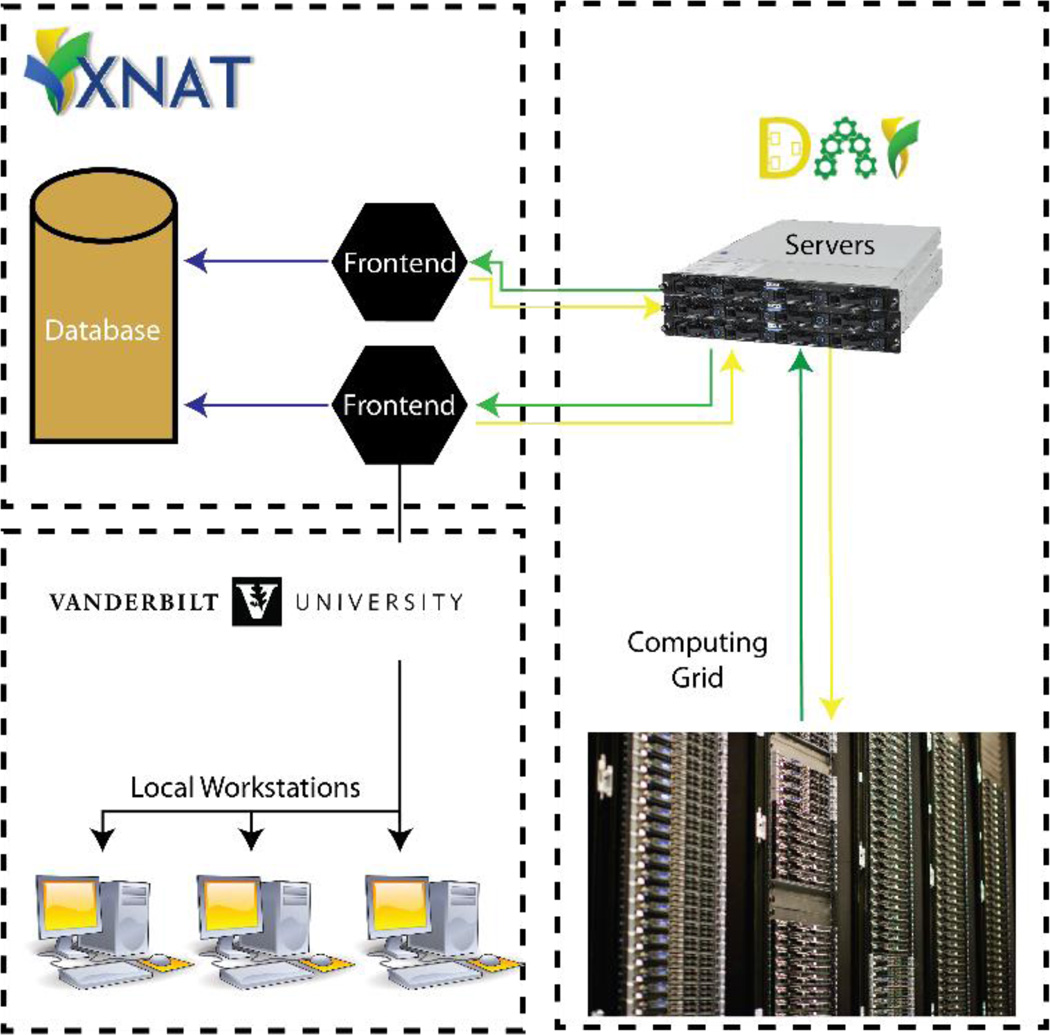
Integration of DAX pipeline with XNAT
by Dipankar Bachar, David Meunier & Aissam Rahmani
External links : Reference :
DAX - The Next Generation: Towards One Million Processes on Commodity Hardware, Stephen M Damon, Brian D Boyd , Andrew J Plassard , Warren Taylor , Bennett A Landman, PMID: 28919661 PMCID: PMC5596878 DOI: 10.1117/12.2254371 (link)
The goals of this BrainHack project are :
- to discuss the working of DAX pipeline and if possible with a live demo
- to understand the working of Spyder in DAX, how to configure it
- to test the DAX command lines
- to try to solve the technical challenges of installations
- to understand/learn the DAX processors
- to understand/learn how to write the YAML scripts
- to learn how to prepare a YAML script for DAX processor in RedCAP.
Required skills
This is a cross-language project. Experience with super computers is also welcome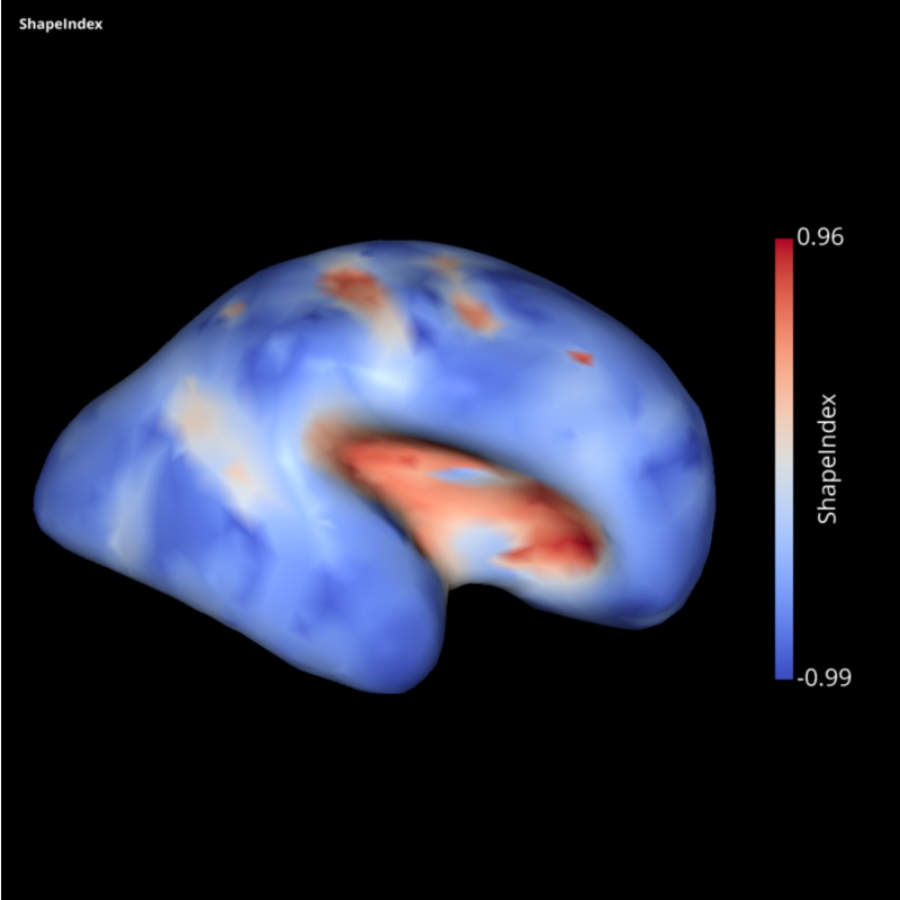
Development of the SLAM -Surface anaLysis And Modeling- python package
by Guillaume Auzias & the MeCA team
Have a look at the examples on the documentation website.
During this brainhack, our objectives are:
- to add as a new feature the algorithms for computing surface profiling as described in Li, K., Guo, L., Li, G., Nie, J., Faraco, C., Cui, G., Zhao, Q., Miller, L.S. and Liu, T., 2010. Gyral folding pattern analysis via surface profiling. NeuroImage, 52(4), pp.1202-1214. https://doi.org/10.1016/j.neuroimage.2010.04.263
- to improve the documentation with new examples to enrich the gallery, which helps a lot potential new users
- to switch the example codes from python script to Jupiter notebooks
- to further improve code quality with new unitest and potential speed-up of specific pieces of code such as for instance the computation of the curvature
- to help potential users to get familiar with this python package
Required skills
Minimal skills in python coding and github are required, but various levels of expertise are welcome since some of our objectives can be addressed with limited coding effort such as augmenting the documentation.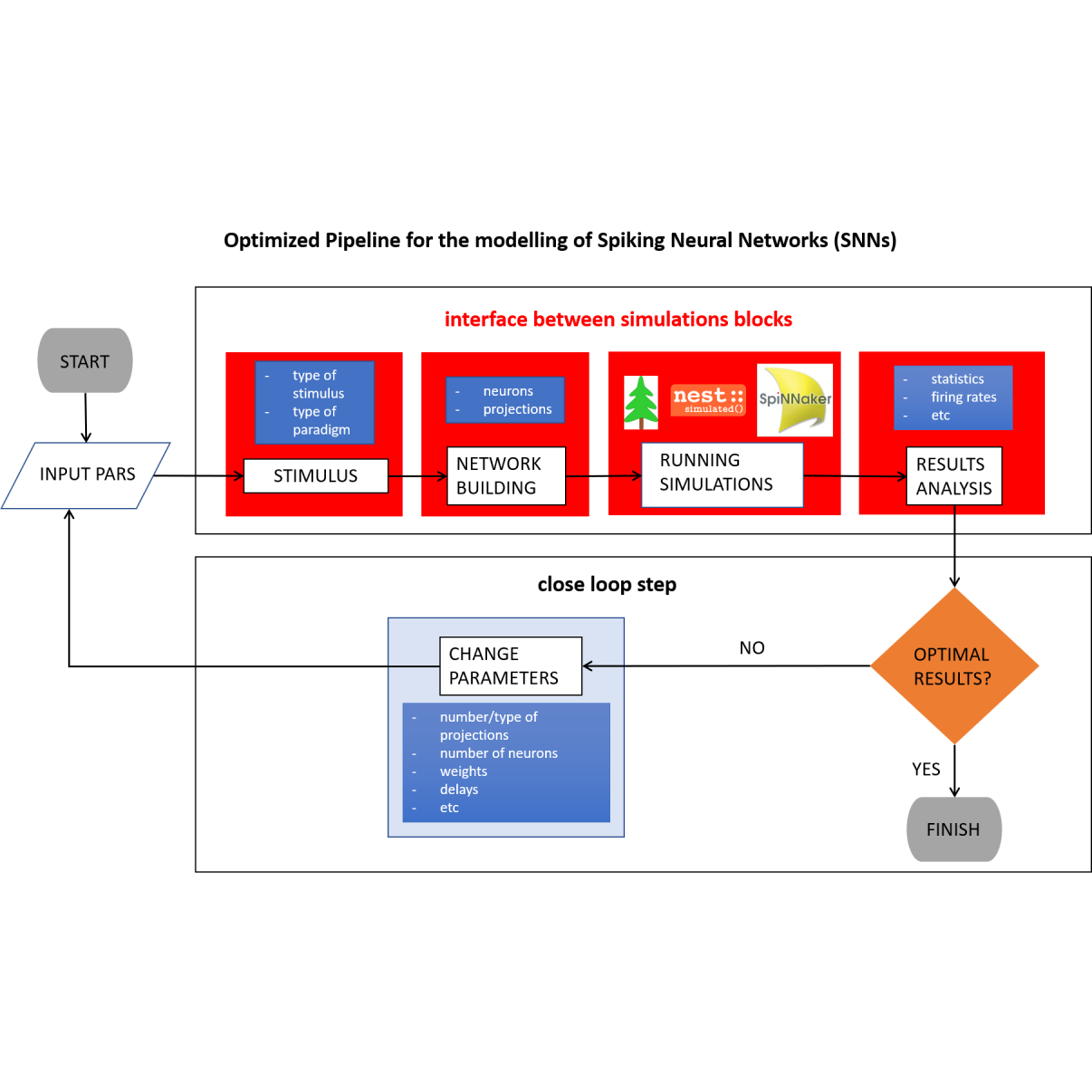
Optimized Pipeline for the modelling of Spiking Neural Networks (SNNs)
by Alberto Vergani, Laurent Perrinet & Julia Sprenger
- setting up the network
- running the simulation
- analyzing the results
Ressources :
- https://github.com/NeuralEnsemble/PyNN
- https://github.com/nest/nest-simulator
- http://spinnakermanchester.github.io/
- Working goals : handle the interface between simulations blocks (network building, running simulations, results analysis)
- Perspective goal : thinking about closing the loop by optimizing the network structure based on the output of the analysis.
Required skills
Skills with standard scientific python librairies are requiredTeam

Sylvain Takerkart
Research Engineer

Guillaume Auzias
Researcher

David Meunier
Research Engineer

Dipankar Bachar
Research Engineer

Julia Sprenger
Engineer

Martin Szinte
Postdoc

Ruggero Basanisi
PhD Student

Etienne Combrisson
Postdoc




Social network