Register
To register to the event, please click on the button below to fill the form. The deadline for registration is Sunday 28 November.
Registration is now closed.
For late registration, you should anyway fill the form (press on the grey 'REGISTER' button) and send an email to the organization team.
Lunch and social event are not guaranteed for late registrations.
Program
Please find bellow the program of the Brainhack event (timezone Paris UTC+1). If you want to add the following schedule to your own personal Google agenda, click on the "Save the dates" button and then click on the "+ Google agenda" located at the bottom right of your screen.
6th December
| 09h00-10h15 | Introduction to BrainHack Marseille 2021
|
|---|---|
| 10h15-12h30 |
Participants can chose to attend one of the two parallel sessions:
Parallel session 1: Hacking Parallel session 2: Python for non-programmers, a tutorial by Julia Sprenger |
| 12h30-14h00 | Break |
| 14h00-16h30 | Hacking |
| 16h30-17h30 | Unconference - Remi Gau, Marie-Hélène Bourget, Martin Szinte & Sylvain Takerkart
BIDS and its recent extensions |
7th December
| 09h00-11h00 | Hacking |
|---|---|
| 11h00-12h30 | Marseille-Lyon exchange session: software demos and discussion |
| 12h30-14h00 | Break |
| 14h00-17h00 | Hacking |
| 17h00-20h00 | BHM social event! |
8th December
| 09h00-11h00 | Hacking |
|---|---|
| 11h0-12h00 | Unconference - Paola Mengotti
Diversity in Academia: The Women in Neuroscience Repository (WiNRepo) |
| 12h00-14h00 | Break |
| 14h00-15h00 | Hacking |
| 15h00-16h00 | Unconference - Greg Operto
Management and Quality Control of Large Neuroimaging Datasets: Developments From the Barcelonaβeta Brain Research Center, (Huguet et al. Frontiers in Neuroscience 2021) |
| 16h00-17h00 | Wrapping up about the projects with everybody |
Projects
Here you can find all the informations about the event projects.
If you want to submit a project you should follow the link, fill the form, and open a github issue. Projects can be anything you'd like to work on during the event with other people (coding, discussing a procedure with coworkers, brainstorming about a new idea), as long as you're ready to minimally organize this!
EyetrackPrep: an automatic tool to pre-process eye-tracking data (fixation, blinks, saccades, micro-saccade, pursuit, pupillometry)
by Martin Szinte, Anna Montagnini & Guillaume Masson
EyeTrackPrep is meant in the future to be used by all researchers using eye-tracking data. It aims at simplifying and standardizing this domain to improve research reproducibility as well as the share of collected data.
Eye-tracking community lack such a tool even if a need for it is clear. Although this project might take time until a functional software is produced, it will modify for long term the way cognitive neuroscientists work and generate open science of high quality data akin what happened with the development of BIDS-apps from the neuroimaging community.
The following points are going to be addressed in this project:
- Discussions about the tool, its goals, what it should and what it should not include
- Modeling of eye movement metrics (e.g. saccade detection from eye-traking position time series)
- Visualization for data quality check
Required skills
This is a python project, a mix of coding and non-coding skills are required:
Integration of the single-trial time-resolved spectral connectivity (coherence; PLV) in Frites
by Vinicius Lima & Etienne Combrisson
Additionally, apart from estimating those metrics in a time-resolved manner, in order to be able to relate the dynamics of the phase-coupling and task-related behavioral events, it is also relevant to assess the dFC at single-trial level, hence, avoiding averaging out non-phase-locked bursts of synchronization that are present in the dFC and may correspond to brain states relevant to determining, for instance, whether the information is being encoded during cognitive tasks by the coordinated activity of multiple cortical areas.
Currently, xfrites - the testing repository associated to Frites - has a function that estimates dFC in terms of the aforementioned metrics. For the present project, we aim to integrate it with Frites and, more specifically, we aim to improve the documentation of the function, refine the current implementation and include code for unit testing. Other goals are to implement notebooks with examples that allow the user to have a better understanding of how to first, set the parameters to estimate the spectral connectivity and seconds, to interpret the metric's outcome, what are its advantages and drawbacks.
Keywords: communication through coherence; spectral analysis; wavelet coherence; dynamic functional connectivity.
The following points are going to be addressed in this project:
- Describe the methods to other participants
- Refine the method implementation to estimate spectral connectivity present in xfrites
- Create the documentation for the method
- Write smoke and functional unit tests
- Create examples illustrating the purpose of the single-trial coherence / PLV
Required skills
This is a python project, the following skills are highly recommended:
BIDS-ephys: let's BIDSify your animal electrophysiology data
by Sylvain Takerkart & Julia Sprenger (coordinators of the BIDS-extension proposal for animal electrophysiology)
Other members of the INCF working group on Neuroscience Data Structure
Our working document that describes the data organization for animal ephys data is available here. It is now ready to be used, and we propose here that electrophysiologists can attempt applying this organization to their data, with the help of the community.
The following points are going to be addressed in this project:
- Have as many electrophysiologists as possible convert their data to the proposed BIDS-ephys organization
- Identify potential problems in the specifications, solve them
- Make progress on the tools required to facilitate this conversion (some of them being present here)
Required skills
This is a project where coding is not required. If you know or have electrophysiology data recorded in animal models, come over and tell us about it! Some other project participants might do some coding, in Python. Appreciated skills:
BrainSpread: Misfolded proteins spreading
by Alessandro Crimi, Luca Gherardini & Aleksandra Pestka
In the project, DWI data collected from Alzheimer's patients is used. The data flow includes: preprocessing, tractography generation, connectivity matrix calculation and connectivity graph visualization. The next step is to simulate a spreading of misfolded proteins based on generated connectivity matrix and the location of seeds for Alzheimer's beta-amyloid in the brain.
The following points are going to be addressed in this project:
- Discuss and code the simulation methods (heat kernel based diffusion, epidemic simulation model etc.).
- Discuss the problem of a connectome structure change in time caused by neurodegeneration
Required skills
This project requires a mix of coding and non-coding skills: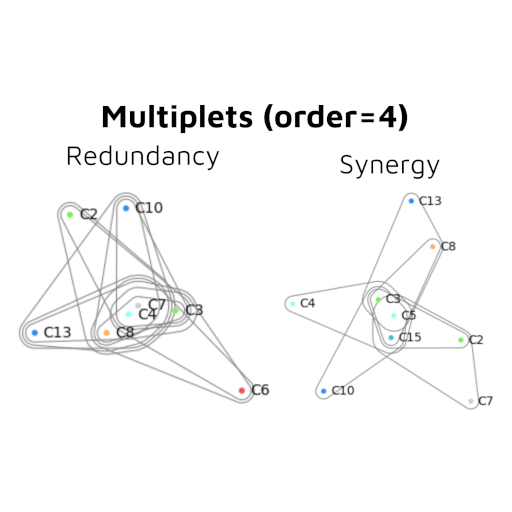
Going beyond pairwise interactions by digging into Higher-Order Interactions
by Etienne Combrisson, Andrea Brovelli & Daniele Marinazzo
Since (d)OInfo frameworks are recent, the math underneath are quite new and we are not necessary familiar with it. The overall goal of this project is to understand the methods by looking at the reference papers and the Python / Matlab implementations of both (d)OInfo.
The following points are going to be addressed in this project:
- Go through the reference papers (i.e. Rosas et al. 2019 and Stramaglia et al. 2021) to build an intuition of the math undergoing the HOI
- Go through the Python toolbox HOI_toolbox to understand what are the input / output types, to identify the main accessible functions such as understanding the internals
- Make the package easy to install, probably clean up so files
- Identify whether there are coding bottlenecks that could be easily solved to speed up computations (soft Numba, multi-core, tensor-computations etc.)
- Implementation of false discovery rate for the significance of the multiplets
- Speed up of the bootstrap
- Include some plotting functions
Required skills
This project requires a mix of coding and non-coding skills:
Macapype: external pipelines
by David Meunier, Kepkee Loh, Julien Sein, Bastien Cagna & Olivier Coulon
Today, macapype is provided as a github repo, a pip install, and a docker/singularity image. The accepted inputs have to be in BIDS format, and macapype pipelines are callable thanks to a command line interface (CLI).
Links
The following points are going to be addressed in this project:
Macapype is mostly oriented to PNH anatomical segmentation. However, many applications can be included starting from a good segmentation quality :
- ACT (anatomically contrained tractography) from diffusion data
- Mesh and surfaces generation that are not pure PNH but that are useful to be linked to macapype segementation
Required skills
This is a Python project, coding skills and imaging knowledge are required: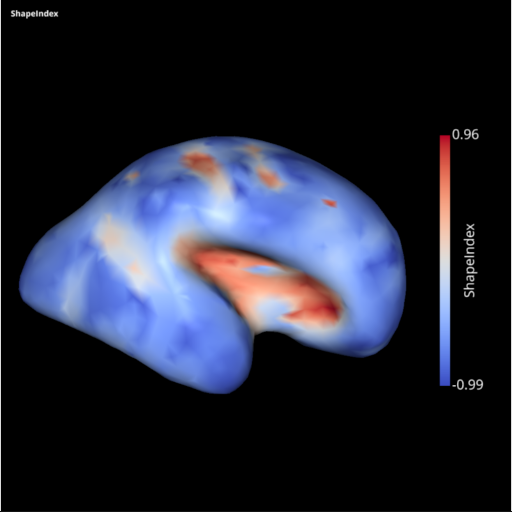
Development of the SLAM -Surface anaLysis And Modeling- python package
by Alexandre Pron, Guillaume Auzias & the MeCA team
Slam is an open source python package dedicated to the representation of neuroanatomical surfaces stemming from MRI data in the form of triangular meshes and to their processing and analysis.
Main features include read/write gifti (and nifti) file format, geodesic distance computation, several implementations of graph Laplacian and Gradient, mesh surgery (boundary identification, large hole closing), several types of mapping between the mesh and a sphere, a disc… Have a look at the examples on the documentation website.
The following points are going to be addressed in this project:
- Help potential users to get familiar with this python package. The first BrainHack day could be considered as a training session for slam users
- Further improve the documentation
- Further improve the code quality of slam 's core functions (code linting, docstrings, refactoring, utils module)
- Further improve code quality with new unitest and potential speed-up of specific pieces of code such as for instance the computation of the curvature
- Brainstorm about slam perimeter
- Design a logo
Required skills
This is a Python project, enthusiasm and coding skills are required: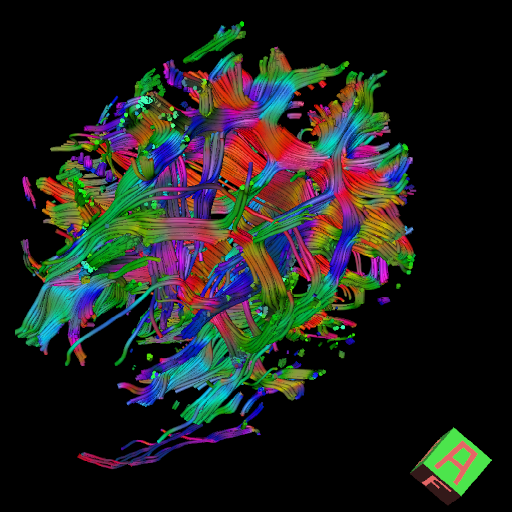
Tools for MRI diffusion in human and NHP
by Julien Sein, David Meunier, Arnaud Le Troter & Hugo Dary
The following points are going to be addressed in this project:
- We aim at sharing expertise, identify specific problems encountered by users, and define quality metrics (quality of orientation vectors, quality check)
- (NHP specific) : We also aim at integrating segmentations computed externally to mrtrix and FSL pipelines, that are non-specific pipelines (i.e. also available for human images)
Required skills
For this project general knowledges about MRI pipelines are recommended:Team

Sylvain Takerkart
Research Engineer

David Meunier
Research Engineer

Dipankar Bachar
Research Engineer

Julia Sprenger
Engineer

Martin Szinte
Researcher

Alexandre Pron
Research Engineer

Simon Moré
Engineer

Ruggero Basanisi
PhD Student

Etienne Combrisson
Postdoc






Social network