Register
To register to the event, please click on the button below to fill the form. The deadline for registration is Friday November the 18th .
Registration is now CLOSED.
Note that there is a limitation of 60 people on site and that the event will be light-hybrid (unconference + some projects).
For late registration, you should anyway fill the form (press on the grey 'REGISTER' button) and send an email to the organization team.
Lunch and social event are not guaranteed for late registrations.
Program
Please find bellow the program of the Brainhack event (timezone Paris UTC+1). If you want to add the following schedule to your own personal Google agenda, click on the "Save the dates" button and then click on the "+ Google agenda" located at the bottom right of your screen.
28th November
| 09h00-10h15 | Introduction to BrainHack Marseille 2022 (In-person/Online)
|
|---|---|
| 10h15-12h30 | Hacking and/or Training:
|
| 12h30-14h00 | Break |
| 14h00-17h30 |
Hacking and/or Training:
|
| 17h30-> | BHM social event! |
29th November
| 09h00-09h30 | Breakfast |
|---|---|
| 09h30-12h30 | Hacking and/or Training:
- FieldTrip by Manuel Mercier - FreeSurfer by Guillaume Auzias - Scikit-Learn by Matthieu Gilson |
| 12h30-14h00 | Break |
| 14h00-15h00 | Hacking |
| 15h00-16h00 | How to archive code with software heritage (In-person/Online) by Sabrina Granger |
| 16h00-17h00 | Hacking and/or Training:
|
| 17h00-17h15 | Break |
| 17h15-18h30 | What Neurosciences In The Anthropocene Era? A participatory workshop (In-person) By Daniele Schön & Julien Lefevre |
30th November
| 09h00-09h30 | Breakfast |
|---|---|
| 09h30-12h30 | Hacking |
| 12h30-14h00 | Break |
| 14h00-16h30 | Hacking |
| 16h30-17h30 | Wrapping up about the projects - closing the BHM 20222 (In-person/Online) |
Projects
Here you can find all the informations about the event projects.
If you want to submit a project you should follow the link, fill the form, and open a github issue. Projects can be anything you'd like to work on during the event with other people (coding, discussing a procedure with coworkers, brainstorming about a new idea), as long as you're ready to minimally organize this!
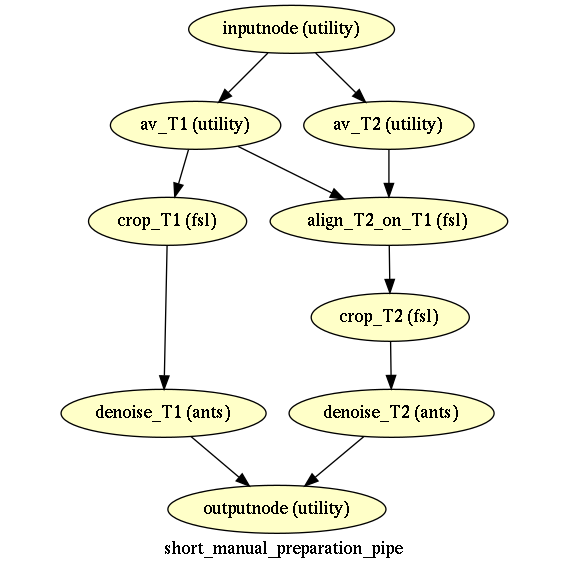
Automatise your processing pipelines with nipype / pydra
Nipype has provided an integrative solution, with a sufficient level of complexity to cover most of the needs for writting pipelines in neuroimaging. It is based on the notion workflows, being an orderd succession of nodes, linking inputs and outputs. Nodes can be user-written function (in python), interfaces with existing softwares (e.g. FSL, AFNI or SPM), or even other user-defined sub-workflows.
Nipype is at the base of many widely used docker images, such as fmriprep and qsiprep. And has been extendend for other applications, such as EEG/MEG processing (ephypype), graph analysis in functional connectivity (graphpype) or non-human primate anatomical MRI segmentation (macapype).
Nipype has now achieved a degree of maturity to have become predominant in the community. But some of the limitations still prevails. It has decided in the last years to rewrite the core engine of nipype, to incorporate new functionnalities, such as runnnig containers as one node. The new implementation will be called pydra, and also still in its infancy, we expect it to become a major standard in the community.
In this project, we propose:
- To give an overview of how nipype works
- To advise you if it is useful for your typical processing
- To help writting specific nodes or workflows dedicated to your processing
For advaced users, We also propose:
- To explain the advances of pydra compaired to nipype
- To write some tools existing in nipype in pydra
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required: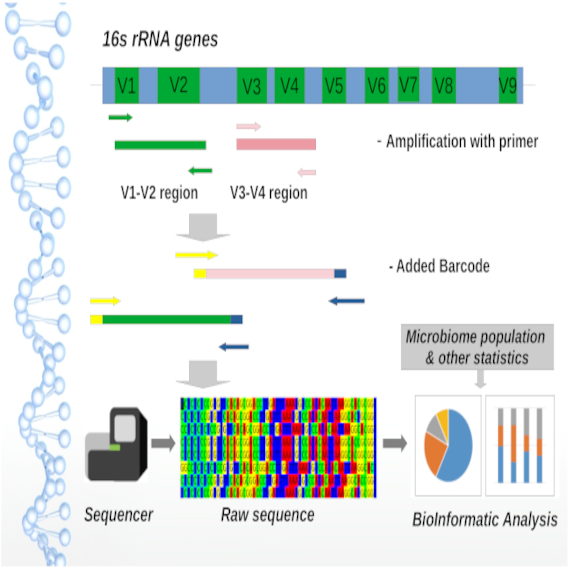
16s Metagenomic analysis of Gut Microbiome
In this project, we propose:
- To go through this [1] article to understand their method of analyzing the gut microbiota
- To download their 16s sequencing data from NCBI (submitted by the authors)
- To extract and filter the data
For advaced users, We also propose:
- To write( Python ) few tools for filtering the sequencing data
- To try to create an Anaconda environment or a Singularity image with the 16s analysis pipeline QIIME2[4]
- To try to implement some of the methods of analysis with QIIME2 (as described in the article)
- To understand the importance of reproducibility of the results
References
[1] The microbial metabolite p-Cresol induces autistic-like behaviors in mice by remodeling the gut microbiota. Patricia Bermudez-Martin et al.
[2] Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson's Disease. Timothy R Sampson et al.
[3] 16S rRNA Gene Sequencing for identification, classification and quantitation of microbes
[4] Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Evan Bolye et al.
[5] Cutadapt
[6] Kraken 2
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required:
LFP and single units decoding in rats performing a distance estimation task
by Fabrizio Capitano, Pierre-Yves Jacob, Celia Laurent & Francesca Sargolini
Here and here you’ll find some of our previous studies to have more insight about the neuroscientific framework of the project.
Goals for the BrainHack:
- Discuss about pros and cons of different decoding methods
- Sort-out the most appropriate one(s) for our project
- Start implementing the analysis on the dataset
- More in general, seeding future collaborations
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required:
SLAM - A pure python package for Surface anaLysis And Modeling
by Guillaume Auzias & Alexandre Pron
Goals for Brainhack:
- To improve the documentation of basic core functions and modules (e.g. the curvature.py module)
- To increase unit test coverage and quality of basic core functions and modules
- To speed-up specific pieces of code such as the computation of the curvatures
- To help potential users to get familiar with this python package depending on their use cases
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required:
Viewing the world in third-person - experimental considerations for a successful study
- I am conceiving an experiment to study the implications of living with a third-person point of view on mental states and self-awareness. For self-awareness implications, see for instance research from Olaf Blanke. I expect positive implications for users, including increased self awareness and increased focus, and interesting implications for users undergoing temporary stress or anxiety. If the issue of learning to associate somatosensory information to the new way of seeing the world can be solved, this could open the door to many more experiments around metacognition, but also around learning involving perception and movement of the body. Inspired by the study of Hubert Dolezal in 1982, using prism glasses to experience the world upside-down.
- Visuo-tactile association in third-person point of view is hard to achieve. The challenge revolves around how to make the brain learn that association. One candidate approach is to start from a first-person point of view and gradually transition to a third-person point of view, using a camera transmitting its video feed to a headset worn by the user. Existing solutions exist for drone piloting (FPV goggles).
- Identify the bottlenecks to come. Propose a design solution to allow for a gradual transition from first-person to third-person point of view. Experiment with FPV goggles and bring them to the Brainhack :) The outcome of this project should be in the form of schematics, prototypes, pseudocode, people to reach out to and a list of resources to take inspiration from.
- https://petapixel.com/2014/07/02/custom-built-oculus-rift-gopro-rig-lets-experience-life-third-person/
- https://www.youtube.com/watch?v=anE3RNf_3s0
Goals for Brainhack:
- The outcome of this project should be in the form of schematics, pseudocode, people to reach out to and a list of resources to take inspiration from.
- Iteration cycles should be as short as possible.
- Testing should ideally happen on-site during the hackathon.
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required: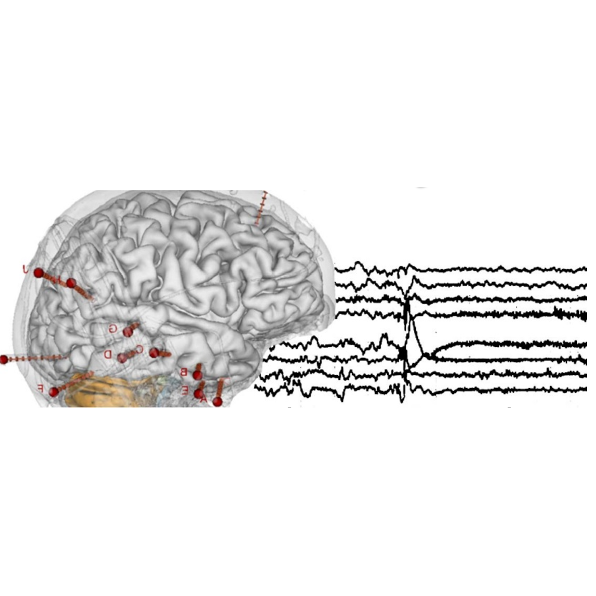
Neural encoding of acoustic features during speech and music perception: traduction of matlab code into python
by Benjamin Morillon, Bruno Giordano, Giorgio Marinato, Nadège Marin & Arnaud Zalta
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required: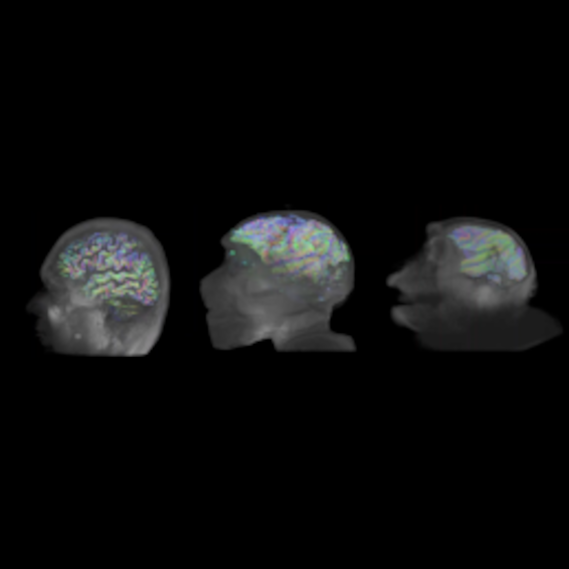
Guidelines for tractography in primates
by Melina Cordeau, David Meunier, Julien Sein & Arnaud Le Troter
Goals for Brainhack:
- Definition of guidelines based on human best practices
- Creation of a parameter file for each primate species
- Implementation of a corresponding Wiki in PRIME-RE platform
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required:
Inferring task-related higher-order interactions from brain network signals
by Etienne Combrisson, Andrea Brovelli, Daniele Marinazzo, Matteo Neri & Ruggero Basanisi
The main goal of this BrainHack is to have a working first version of the task-related HOI:
- Prototype the main function (i.e. define input and output types, write down important internal steps)
- Make it works in the non-dynamic case
- Investigate the use of Jax to speed up computations
- Be able to simulate data with a known amount of redundancy and synergy
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required: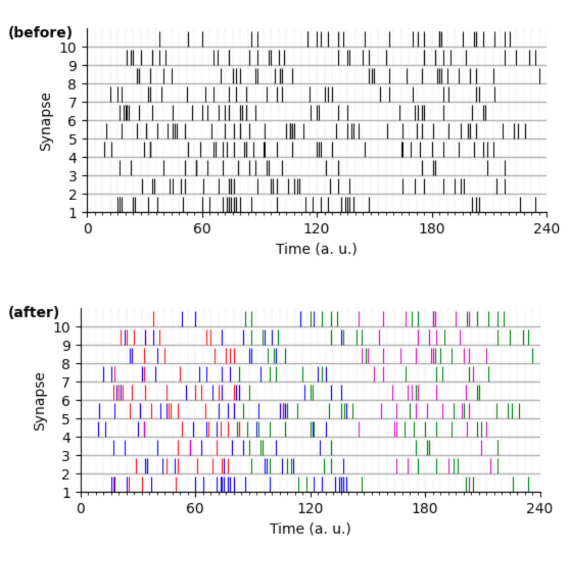
Automatic detection of spiking motifs in neurobiological data
by Matthieu Gilson, Laurent Perrinet, Hugo Ladret & George Abitbol
An implementation could be based on autodifferentiable networks as implemented in Python libraries like pytorch. This framework allows for the tuning of parameters with specific architectures like convolutional layers that can capture various timescales in spike patterns (e.g. latencies) in an automated fashion. Another recent tool based on the estimation of firing probability for a range of latencies has been proposed (Grimaldi ICIP 2022). This will be compared with existing approaches like Elephant’s SPADE or decoding techniques based on computed statistics computed on smoothed spike trains (adapted from time series processing, see (Lawrie, biorxiv).
One part concerns the generation of realistic synthetic data producing spike trains which include spiking motifs with specific latencies or comodulation of firing rate. The goal is to test how these different structures, which rely on specific assumptions about e.g. stationarity or independent firing probability across time, can be captured by different detection methods.
Bring you real data to analyze them! We will also provide data from electrophysiology.
Goals for Brainhack:
- Code to generate various models of synthetic data (time series of spikes/events) with embedded patterns
- Knowledge in signal processing & high-order statistics (correlation)
- Tool for quantitative comparison of detection methods for correlated patterns
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required:
BIDS, electrophysiological data, open data
by Sylvain Takerkart & Julia Sprenger
Goals for Brainhack:
- Chatting, discussing, making progress!
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required:
PsychophyGit - a fast and flexible tool for online experiments
by Hugo Ladret, Jean-Nicolas Jérémie & Laurent Perrinet
Goals for Brainhack:
- Develop Python code that allows to deploy psychophysical experiments with light requirements
- Develop a server-side interface which parses GitHub repos to get experimental stimuli
- Create simple 2 Alternative Forced Choice tasks
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required:
Brainhack projects database
by Rémi Gau
The brainhack has been running for more than 10 years, yet we do not have single centralised resource to show in a fairly exhaustive manner the diversity of projects that have happened over the past decade.
Having a quick way to create reports about the success of brainhack could make significantly easier for event organizer to look for funding.
Solution
In the past few years more and more events have started listing their projects as github issues.
This now makes it easier to:
- start creating a mini database of:
- brainhack events
- brainhack sites
- brainhack projects
- to create an interactive dashboard to query and create visualizations of that database
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required:
Sulcilab: A collaborative sulci labelling web app
Sulcilab is a web base application to mannually labelize sulcal graphs (outputs of BrainVISA).
I propose this project with 2 goals:
- Get feedback from beta users
- Propose to develop a javascript based package to provide a 2D/3D viewer for neuroimaging
- Ask me to create an account for you and then go to http://babalab.fr:3000 to test the app.
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required: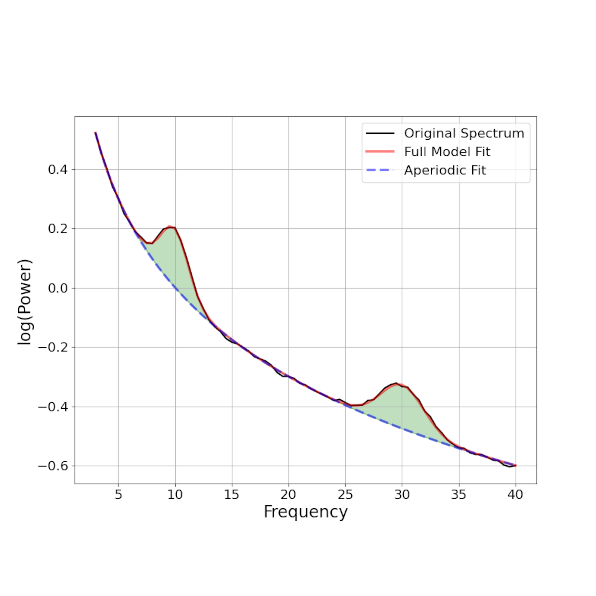
Removing pink noise (1/f) in LFP across different electrophysiological recording systems
by Laura López Galdo, Shrabasti Jana, Camila Losada, Hasnae Agouram, Cléo Schoeffel & Nilanjana Nandi
We will try to remove the aperiodic noise found in the spectrum of the LFP data using the fooof module (https://github.com/fooof-tools/fooof). We will parametrize our signals, the aperiodic and periodic components and make some comparison across the different frequency bands.
We have data from EEG human recordings, LFP monkey utah array recordings and monkey laminar intracranial recordings across different areas. We want to see the effect of pink noise in each of the different setups and try to clean the signal.
Some related literature can be found in the following links:
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required:
Decoding the bulk signal from fiber photometry during rat behavior
by Maya Williams, Mickael Degoult & Christelle Baunez
I am collecting calcium signals from Infalimbic, Prelimbic, and Anterior cingulate projection terminals at the subthalamic nucleus (STN) using fiber photometery. The goal is to look at the signal from these regions during sucrose and cocaine self administration, to discover the role of these pathways in food vs cocaine intake. In addition, we would focus on discrete behavior during the trails ( lever pressing, reward delivery, error responses) to study the signals of these pathways during both sucrose and cocaine intake.
It will be the first time the hyperdirect pathway is studied in such detail, looking at the mulitpule cortical projection the STN and comparing their roles in food vs cocaine rewards. From perovious work in the lab, we know that STN lesions have opposite effects on cocaine and food intake, increasing food intake while decreasing cocaine intake. This study will explore the much less studies hyperdirect pathway and its role in addiction like behaviors so that the STN can be consided a target for theraputic treatment in addiction, without harming a persons natural motivations for food.
I have collected data from sucrose taking rats, and addapted an open souce code to fit my data. I next need help on further analysis, combinding results into groups, and doing statistical analysis.
Some codes I have used or wish to try:
Required skills
This is a multimodal project, a mix of coding and non-coding skills are required:Team

Sylvain Takerkart
Research Engineer

David Meunier
Research Engineer

Dipankar Bachar
Research Engineer

Julia Sprenger
Engineer

Melina Cordeau
PhD student

Alexandre Pron
Research Engineer

Simon Moré
Engineer

Ruggero Basanisi
Postdoc

Etienne Combrisson
Postdoc

Guillaume Auzias
Researcher
Matthieu Gilson
Junior Professor

Manuel Mercier
Research Engineer

Arnaud Le Troter
Research Engineer













Social network